参考库是什么?
在运行GATK等工作流时,需要使用一些参考基因组文件及其索引文件、数据库文件及其索引文件。如果这些文件都作为输入项文件,一方面用户需要在做工具的时候需要设置多个输入项、运行工作流的时候需要填写较多的输入文件。另一方面这些文件比较大,运算前需要下载到容器里,需要耗费较多时间。
在针对上述场景,GeneDock 提供了公共参考库文件和自定义参考库功能。
GeneDock公共参考库
GeneDock提供公共的参考文件,该文件在容器里的路径为:/rdata/genedock/,用户可在工具里使用这些文件,例如 ls /rdata/genedock/hg19_broad/ucsc.hg19.fasta。
GeneDock提供hg19、b37 和 hg38三类参考文件,用户既可以使用自己的参考文件也可以使用GeneDock的参考文件。
hg19参考文件列表如下
hg19_broad/1000G_omni2.5.hg19.sites.vcf
hg19_broad/1000G_omni2.5.hg19.sites.vcf.idx
hg19_broad/1000G_phase1.indels.hg19.sites.vcf
hg19_broad/1000G_phase1.indels.hg19.sites.vcf.idx
hg19_broad/1000G_phase1.snps.high_confidence.hg19.sites.vcf
hg19_broad/1000G_phase1.snps.high_confidence.hg19.sites.vcf.idx
hg19_broad/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf
hg19_broad/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.idx
hg19_broad/dbsnp_138.hg19.excluding_sites_after_129.vcf
hg19_broad/dbsnp_138.hg19.excluding_sites_after_129.vcf.idx
hg19_broad/dbsnp_138.hg19.vcf
hg19_broad/dbsnp_138.hg19.vcf.idx
hg19_broad/hapmap_3.3.hg19.sites.vcf
hg19_broad/hapmap_3.3.hg19.sites.vcf.idx
hg19_broad/ucsc.hg19.dict
hg19_broad/ucsc.hg19.fasta
hg19_broad/ucsc.hg19.fasta.amb
hg19_broad/ucsc.hg19.fasta.ann
hg19_broad/ucsc.hg19.fasta.bwt
hg19_broad/ucsc.hg19.fasta.fai
hg19_broad/ucsc.hg19.fasta.nhr
hg19_broad/ucsc.hg19.fasta.nin
hg19_broad/ucsc.hg19.fasta.nsd
hg19_broad/ucsc.hg19.fasta.nsi
hg19_broad/ucsc.hg19.fasta.nsq
hg19_broad/ucsc.hg19.fasta.pac
hg19_broad/ucsc.hg19.fasta.sa
b37参考文件列表如下
b37_broad/1000G_omni2.5.b37.vcf
b37_broad/1000G_omni2.5.b37.vcf.idx
b37_broad/1000G_phase1.indels.b37.vcf
b37_broad/1000G_phase1.indels.b37.vcf.idx
b37_broad/1000G_phase1.snps.high_confidence.b37.vcf
b37_broad/1000G_phase1.snps.high_confidence.b37.vcf.idx
b37_broad/1000G_phase3_v4_20130502.sites.vcf
b37_broad/1000G_phase3_v4_20130502.sites.vcf.idx
b37_broad/Broad.human.exome.b37.interval_list
b37_broad/Mills_and_1000G_gold_standard.indels.b37.vcf
b37_broad/Mills_and_1000G_gold_standard.indels.b37.vcf.idx
b37_broad/dbsnp_138.b37.excluding_sites_after_129.vcf
b37_broad/dbsnp_138.b37.excluding_sites_after_129.vcf.idx
b37_broad/dbsnp_138.b37.vcf
b37_broad/dbsnp_138.b37.vcf.idx
b37_broad/hapmap_3.3.b37.vcf
b37_broad/hapmap_3.3.b37.vcf.idx
b37_broad/hs37d5/hs37d5.dict
b37_broad/hs37d5/hs37d5.fasta
b37_broad/hs37d5/hs37d5.fasta.amb
b37_broad/hs37d5/hs37d5.fasta.ann
b37_broad/hs37d5/hs37d5.fasta.bwt
b37_broad/hs37d5/hs37d5.fasta.fai
b37_broad/hs37d5/hs37d5.fasta.pac
b37_broad/hs37d5/hs37d5.fasta.sa
b37_broad/human_g1k_v37.dict
b37_broad/human_g1k_v37.fasta
b37_broad/human_g1k_v37.fasta.amb
b37_broad/human_g1k_v37.fasta.ann
b37_broad/human_g1k_v37.fasta.bwt
b37_broad/human_g1k_v37.fasta.fai
b37_broad/human_g1k_v37.fasta.nhr
b37_broad/human_g1k_v37.fasta.nin
b37_broad/human_g1k_v37.fasta.nsq
b37_broad/human_g1k_v37.fasta.pac
b37_broad/human_g1k_v37.fasta.sa
hg38参考文件列表如下
hg38_broad/1000G_omni2.5.hg38.vcf
hg38_broad/1000G_omni2.5.hg38.vcf.idx
hg38_broad/1000G_phase1.snps.high_confidence.hg38.vcf
hg38_broad/1000G_phase1.snps.high_confidence.hg38.vcf.idx
hg38_broad/Mills_and_1000G_gold_standard.indels.hg38.vcf
hg38_broad/Mills_and_1000G_gold_standard.indels.hg38.vcf.idx
hg38_broad/dbsnp_138.hg38.vcf
hg38_broad/dbsnp_138.hg38.vcf.idx
hg38_broad/hapmap_3.3.hg38.vcf
hg38_broad/hapmap_3.3.hg38.vcf.idx
hg38_broad/hg38.chrom.sizes
hg38_broad/hg38.dict
hg38_broad/hg38.fasta
hg38_broad/hg38.fasta.amb
hg38_broad/hg38.fasta.ann
hg38_broad/hg38.fasta.bwt
hg38_broad/hg38.fasta.fai
hg38_broad/hg38.fasta.pac
hg38_broad/hg38.fasta.sa
自定义参考库
有时,用户需要使用的参考库文件不在GeneDock提供的公共参考库。针对这一场景,GeneDock提供了自定义参考库功能,用户自己管理参考库文件。
- 创建 ref目录
使用ref盘之前,需要在账号根目录下创建/ref/。如果已经创建请跳过此步骤。
- 上传数据到ref目录
将需要用到的参考库文件上传到ref目录,比如 将 hg19_broad/目录上传到 /ref/目录下。ref目录的结构如下:
ls /ref/hg19_broad/
dbsnp_138.hg19.vcf
dbsnp_138.hg19.excluding_sites_after_129.vcf
Mills_and_1000G_gold_standard.indels.hg19.sites.vcf
ucsc.hg19.fasta.fai
1000G_phase1.snps.high_confidence.hg19.sites.vcf
1000G_phase1.indels.hg19.sites.vcf
1000G_omni2.5.hg19.sites.vcf
hapmap_3.3.hg19.sites.vcf
ucsc.hg19.fasta
ucsc.hg19.dict
- 使用自定义参考库文件
/ref/ 云端目录会挂载到容器的 /genedock/ref/目录。
例如,之前上传的hg19_broad 目录在容器里的路径是:/genedock/ref/hg19_broad/。
在 “编辑工具”- “命令行模板” 的时候,可以在命令行里直接使用 ref盘数据。例如:
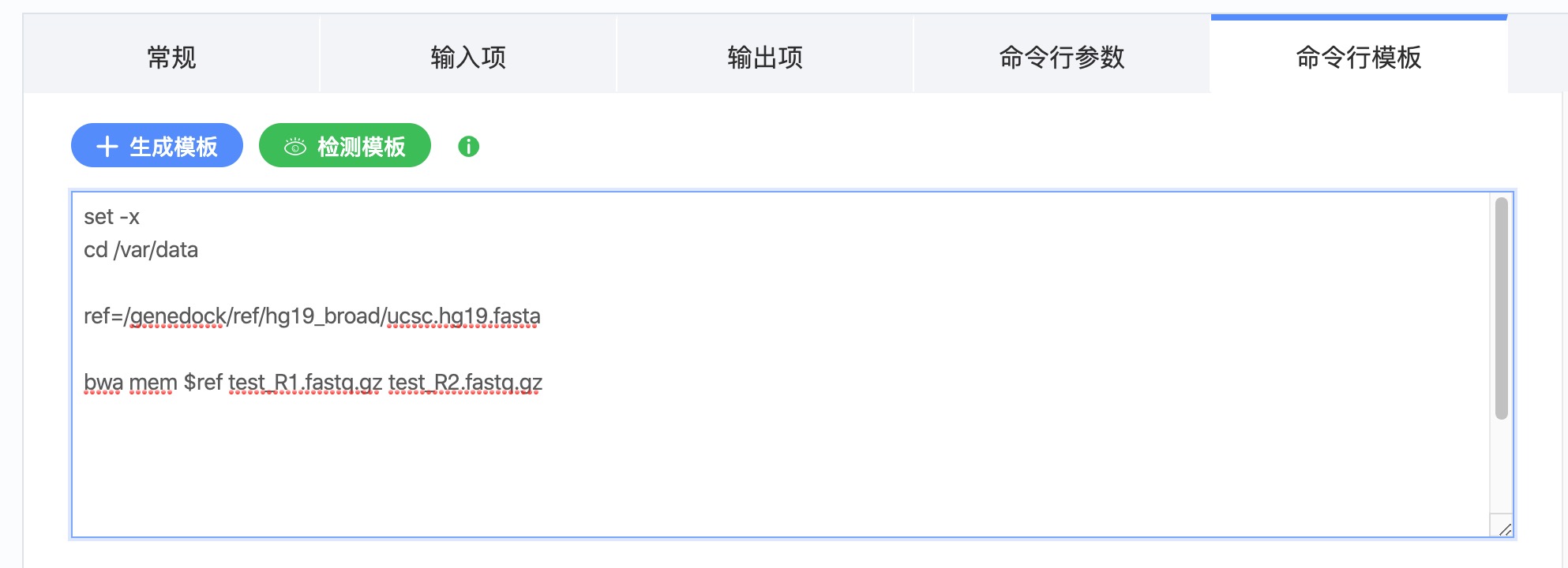
注意事项
请注意,公共参考库的容器路径是
/rdata/genedock/,自定义参考库的容器路径是/ref/genedock/。ref盘的数据安全性
作业里挂载的ref盘数据与作业启动时<account>:/ref/ 目录的数据一致。每个账号挂载的是该工具账号下的ref数据。
- 挂载ref盘的时候会挂载那些数据?
会将 <account>:/ref/ 目录下的所有数据挂载到 /genedock/ref/目录下。
- ref盘也分域
在北京域启动的作业挂载的是北京域的 <account>:/ref/,同理深圳域。